Contents
% EKF vs. UKF % 2014. 9. 24 % refopen.blogspot.com % This example intuitively shows that the difference between EKF and UKF % when state transition is non-linear function. function filterComparison()
Create real data
Create some real data points(1000points)
sigmax = 5; sigmay = 3; rx = randn(1000, 1)*sigmax; ry = randn(1000, 1)*sigmay; % real data points and MLE parameter estimation figure(001); plot(rx, ry, 'r.'); xlabel('x'); ylabel('y'); axis([-15 15 -15 15]); [x, P] = MLEnPlot(rx, ry);


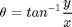
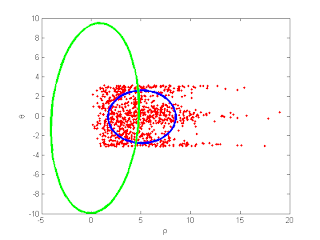
figure(002); rho = sqrt(rx.^2 + ry.^2); theta = atan2(ry, rx); plot(rho, theta, 'r.'); xlabel('\rho'); ylabel('\theta'); MLEnPlot(rho, theta); fprintf('press any key to continue...\n'); pause;
press any key to continue...

Extended Kalman Filter
Find out jacobian matrix when it transformed by non-linear state transition matrix.![$$J = \left[\begin{array}{cc} \frac{\partial r}{\partial x} & \frac{\partial r}
{\partial y} \\ \frac{\partial \theta}{\partial x} & \frac{\partial
\theta}{\partial y} \end{array}\right]$$](https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEidMCLowrdRWWA3hvetRFLeI2MiKMm6yPLdtgUc5jibvGy3Cnv2T9MIS8YeuMHfInE8ZVA5caXIZgUHkm2s1m-7OPQx4f9JOVr2qZYdNN3FisbeLw-eHdIqkqzTzC0Oh6fT47y7KeSLSdXq/s1600/filterComparison_eq58272.png)
Now, prediction of covariance matrix at the pridiction step

q = x(1).^2 + x(2).^2; J = [x(1)/sqrt(q) x(2)/sqrt(q); -x(2)/q x(1)/q]; xekf = [sqrt(x(1).^2 + x(2).^2) atan2(x(2), x(1))]; Pekf = J*P*J'; % draw ellipse e = make_covariance_ellipses(xekf, Pekf); hold on; plot(e(1, :), e(2, :), 'g-' , 'LineWidth', 3); hold off; fprintf('press any key to continue...\n'); pause;
press any key to continue...
Unscented Kalman Filter
Set up some valuesD = length(x); % dimension
N = D*2 + 1;
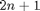 samples are should be generated.
samples are should be generated.scale = 3;
kappa = scale-D;
% Create unscented sample points
Ps = chol(P)' * sqrt(scale);
ss = [x', repvec(x',D)+Ps, repvec(x',D)-Ps];

rho = sqrt(ss(1, :).^2 + ss(2, :).^2); theta = atan2(ss(2, :), ss(1, :)); x = [rho; theta]; idx = 2:N; xukf = (2*kappa*x(:,1) + sum(x(:,idx),2)) / (2*scale); diff = x - repvec(xukf,N); % weighted sample cov. Pukf = (2*kappa*diff(:,1)*diff(:,1)' + diff(:,idx)*diff(:,idx)') / (2*scale); % draw ellipse e = make_covariance_ellipses(xukf, Pukf); hold on; plot(e(1, :), e(2, :), 'k-' , 'LineWidth', 3); hold off;

end function [x, P] = MLEnPlot(x, y) phatx = mle(x); phaty = mle(y); x = [phatx(1) phaty(1)]; P = [phatx(2) 0; 0 phaty(2)]; e = make_covariance_ellipses(x, P); hold on; plot(e(1, :), e(2, :), 'b-' , 'LineWidth', 3); hold off; end

from timbailey's matlab utility
function p= make_covariance_ellipses(x,P) % compute ellipses for plotting state covariances N= 60; inc= 2*pi/N; phi= 0:inc:2*pi; lenx= length(x); lenf= (lenx-3)/2; p= zeros (2,(lenf+1)*(N+2)); ii=1:N+2; p(:,ii)= make_ellipse(x(1:2), P(1:2,1:2), 2, phi); ctr= N+3; for i=1:lenf ii= ctr:(ctr+N+1); jj= 2+2*i; jj= jj:jj+1; p(:,ii)= make_ellipse(x(jj), P(jj,jj), 2, phi); ctr= ctr+N+2; end end % % function p= make_ellipse(x,P,s, phi) % make a single 2-D ellipse of s-sigmas over phi angle intervals r= sqrtm(P); a= s*r*[cos(phi); sin(phi)]; p(2,:)= [a(2,:)+x(2) NaN]; p(1,:)= [a(1,:)+x(1) NaN]; end function x = repvec(x,N) x = x(:, ones(1,N)); end
댓글 없음:
댓글 쓰기